 |
|
Spotlight Our widgets for functional genomics use Orange, a data mining
and machine learning suite. Orange can be accessed through scripting in
Python,
or by visual programming in Orange Canvas. |
| FRI > Biolab > Supplements > Microarray Data Mining with Visual Programming > S. cerevisiae Example > Schema | |
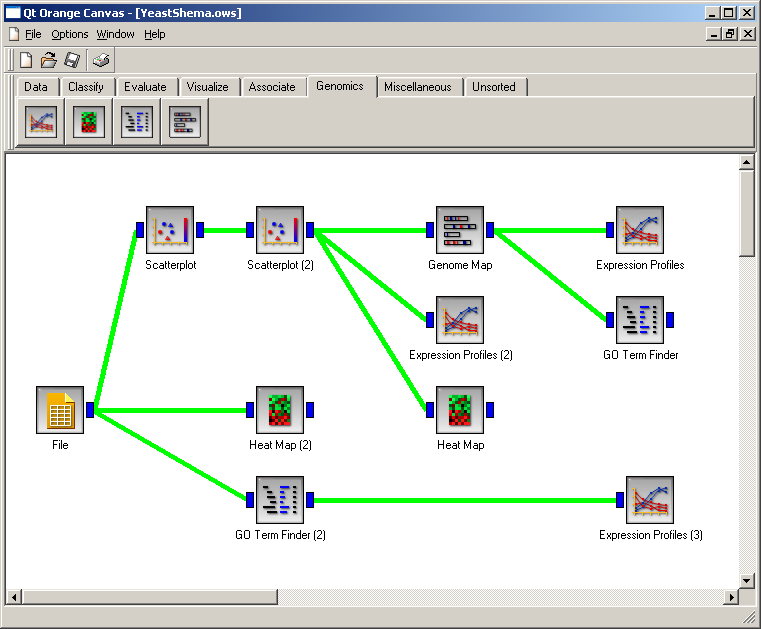
SchemaWe have designed a schema in which two "Scatter Plot" widgets are first used to isolate the genes whose expression is specifically induced in the M/G1 phase of the cell cycle, as shown in "Expression Profiles (2)" and "Heat Map" widgets. "Genome Map" widget is used to demonstrate the adjacent positioning of coexpressed genes, as described in Cho et al. (1998). "GO Term Finder" widget is used to show that two adjacent coexpressed genes, CDC46 and PIG1 are functionally unrelated, and the "GO Term Finder (2)" widget is used to show the GO biological_process categories of the 800 input genes. Clicking on a widget icon in the schema displays a snapshot of the widget during analysis. Please wait for the "GO Term Finder (2)" widget to finish before exploring data in other widgets.
|
|