 |
|
Spotlight Our widgets for functional genomics use Orange, a data mining
and machine learning suite. Orange can be accessed through scripting in
Python,
or by visual programming in Orange Canvas. |
| FRI > Biolab > Supplements > Microarray Data Mining with Visual Programming > D. discoideum Example > GO Term Finder widget | |
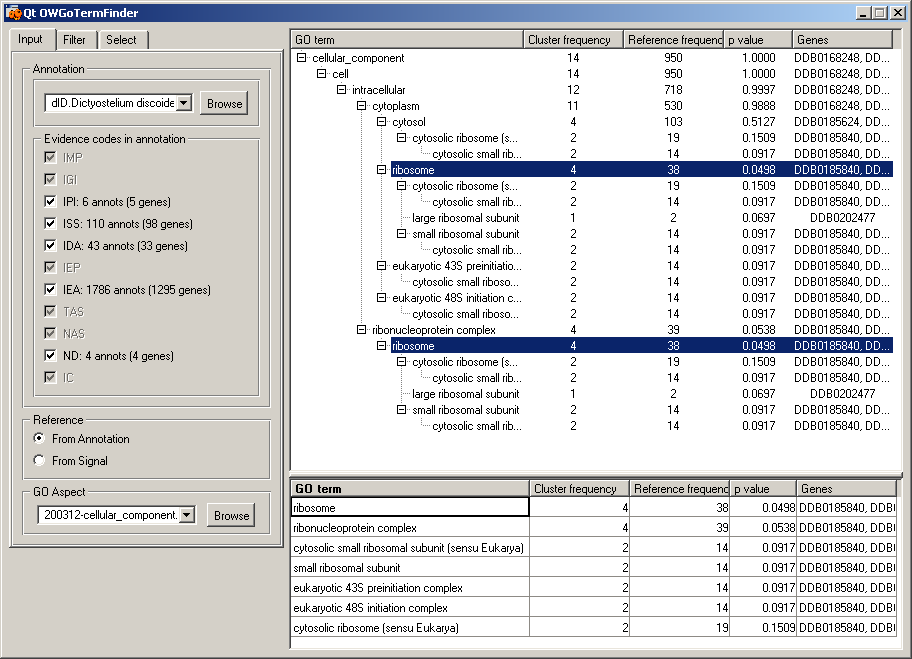
GO Term Finder widgetInput to this widget are genes from cluster "1" that were selected in the "Heat Map" widget. We have used the cellular component aspect of the Gene Ontology to find significant GO terms associated with input genes. Ribosome and cytosolic genes appear among the most significant GO terms. This is in concordance with the study (Van Driessche et al., 2002). The GO aspects and the annotation data files to be used are selected in the two pull-down combo boxes (files listed on main page: cellular_component and annotation). GO terms can be filtered on the number of genes in the term and on the calculated p value. We select genes by selecting GO terms. All data associated with the selected genes is sent as a new data token. We have selected the "ribosome" GO term. Data for the four genes (DDB0185840,DDB0185051,...) in sent to the two linked widgets: "Data Table" and "Expression Profiles (2)."
|
|