Experimental analysis
The performance of the proposed algorithm was assessed on different
gene networks that all include
genes which were most differentially expressed in the leukemia gene
expression data set (Golub et al., 1999). The original data set includes
7,074 genes whose expression was measured using DNA microarrays in 72
tissue samples classified either as acute lymphoblastic leukemia (ALL,
48 samples) or acute myeloid leukemia (AML, 25 samples). We selected
1,025 differentially expressed genes according to Student's
t-statistic significantly smaller or larger (p-value
< 0.01) with respect to the null distribution of the statistic. The
null distribution was obtained by randomly permuting the class labels
and calculating the t-statistic for all the genes. In all the networks the genes represented with solid circles were significantly over-expressed in the ALL samples and the
genes shown as empty circles had higher expression in the AML samples. For
additional assistance to the interpreter, the network components
were named with Gene Ontology terms (Ashburner et al., 2000).
Based on the set of differentially expressed genes and different means
to estimate the gene similarity, we have defined five distinct gene
networks.
Network N1
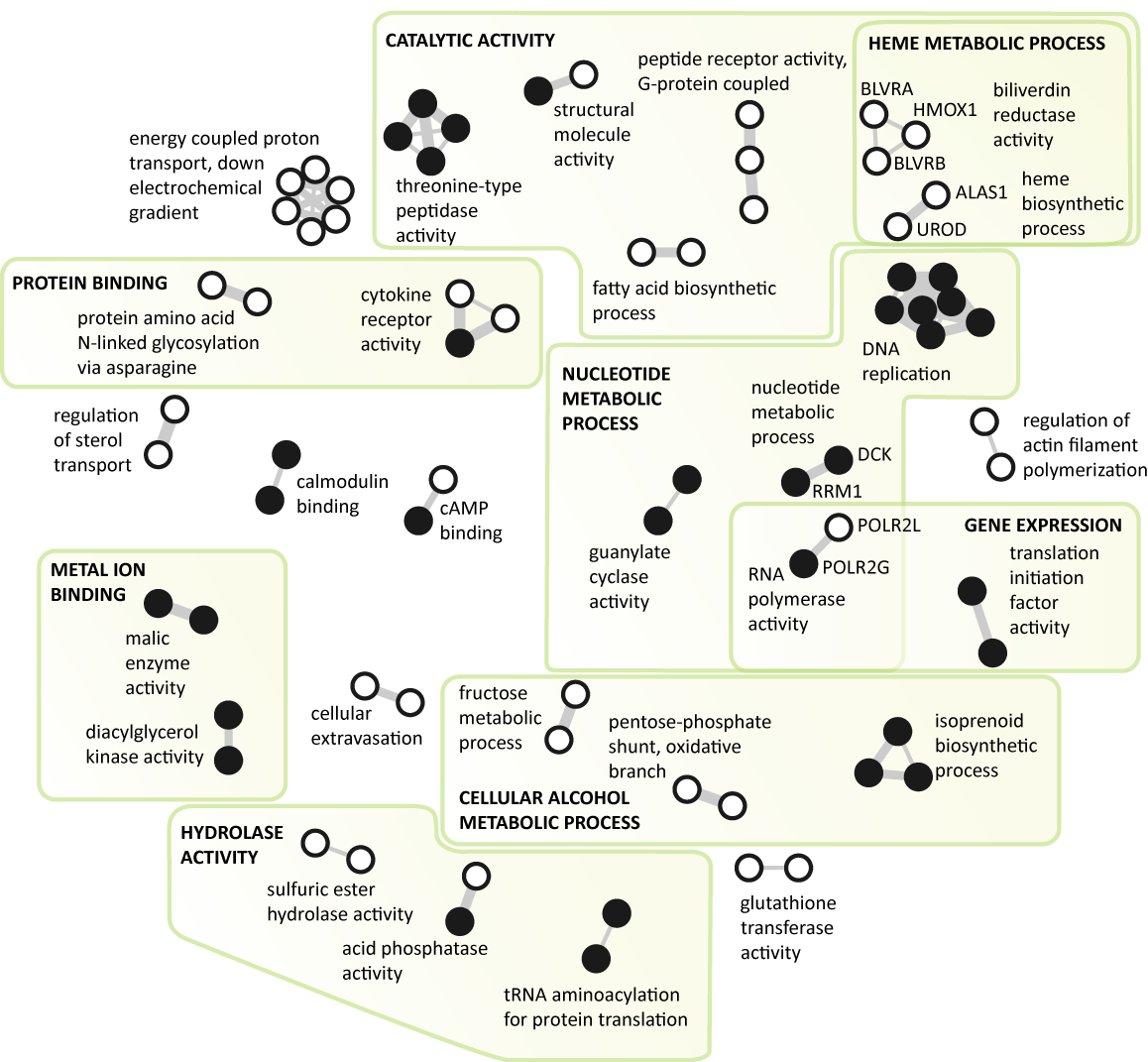
The similarity of the genes relates to their biological functions and was calculated based on their membership in canonical biological pathways using the Jaccard index. The information on the membership of
genes in biological pathways was acquired from the Molecular Signature Database (Subramanian et al., 2005) (C2 collection, canonical pahways). The similarity threshold was set to 0.7 and all the unconnected genes were removed.
The "guanylate cyclase activity", "nucleotide metabolic process",
"RNA polymerase activity", and "DNA replication" components
in N1 all connect genes significantly over-expressed in acute
lympoblastic leukemia. All of these genes have a function in
nucleotide metabolism and DNA biosynthesis. It is well known
that lymphoblastic cells typically have severalfold higher activity of
enzymes responsible for nucleotide metabolism enabling excessive
proliferation of transformed cells (Scholar and Calabresi, 1973).
Moreover, some of the pathways active in nucleotide metabolism,
for example de novo purine synthesis (DNPS), have been recognized
as important targets of antileukemic agents (eg. methotrexate,
mercaptopurine). In combination with other therapeutical agents,
these drugs have improved survival of children with ALL to an
overall cure rate of approximately 80 percent (Pui and Evans,
2006). The N1 network clearly demonstrates this
characteristic of acute lymphoblastic leukemia.
Data files:
distance matrix (tutorial on this data), distance matrix data, network (tutorial on this data), network data
Network N2.1
The similarity between genes as computed by Huttenhower et al., 2009 using the information on all publicly available gene expression and protein interaction data, combined with prior knowledge from the Gene Ontology, KEGG, HPRD and other biological data bases. Similarity scores for the leukemia genes were used to build the N2 network, where only connected genes are shown (genes connected to at least one other gene). The threshold for similarity was set to 0.999.
As in network N1, most of the graph
components connect genes that are over-expressed in one of the
two investigated kinds of leukemia (all genes in the component are
the same color), demonstrating the well known phenomenon that
not only individual genes, but whole processes and pathways are
disrupted in cancer cells (Hanahan and Weinberg, 2000).
Data files:
distance matrix, distance matrix data, network (a), network data (a)
Network N2.2

For this network the same similarity scores and threshold as in N2 were used (the Huttenhower et al., 2009 similarity score). But, differently to N2, N3 also includes unconnected vertices (genes not connected to any other gene), in order to observe the similarity of all the differentially expressed genes.
One can observe that the genes significantly
differentially expressed in the two investigated leukemias cluster
together. The empty circles (AML) are clustered in the right
part of the graph and the solid ones (ALL) in the left part,
again demonstrating that expression changes in cancer tissues are
disrupted on the level of pathways and processes.
Data files:
network (b), network data (b)
Network N3

Protein-protein interaction network: theleukemia genes were connected into the network based on their protein interactions from the MIPS mammalian protein-protein interaction database [19]. In addition, we used the biological function similarity score (described under N1) for placing the interacting protein components based on the similar biological functions of the proteins comprising them.
Several gene products (proteins) that lie close to each other in the FragViz* optimized network are actually in interaction based on a different public repository that stores protein-protein interactions identified by experimental results, the Human Protein Reference Database (HPRD) [25]. For example, the protein Integrin beta 3 (itgb3 ) is, based on the data in HPRD, in inter action with protein Integrin beta 1 (itgb1). Also, proteins Poly A polymerase alpha (papola) and SMAD3 are both in interaction with protein smad2. According to HPRD, protein interactions also exist among proteins in the components il4r-htatip and the near-lying component in the optimized layout.
Data files:
distance matrix, network, network data
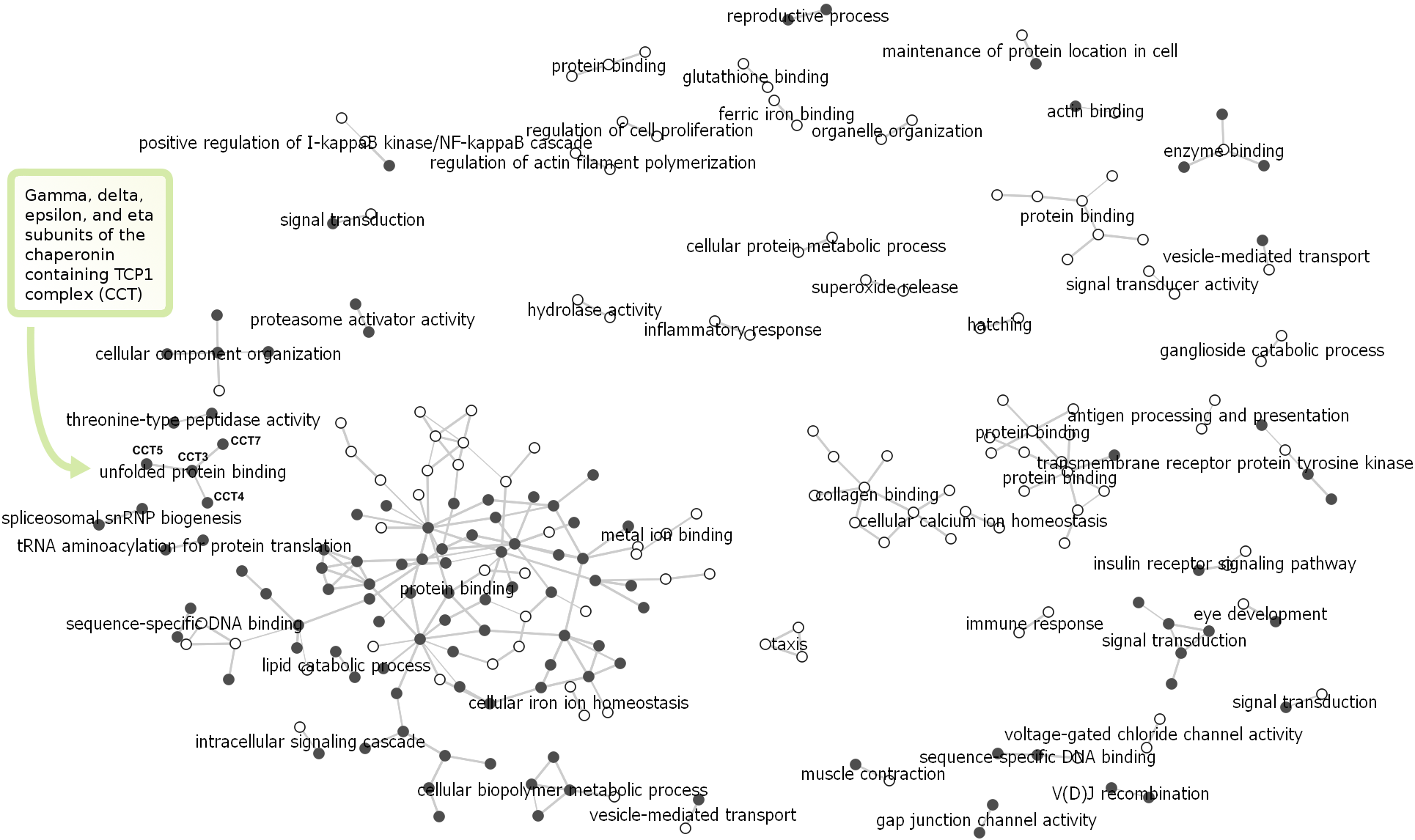
Network N4

The gene similarities are calculated based on common reference in articles form the PubMed using the Jaccard index. Two genes are similar if they are referenced together in several PubMed articles. The similarity threshold was set to 0.5.
Notice that the largest component consists mainly of genes
associated with nucleic acid binding. Names of many other
components in the network are associated with gene expression,
DNA biosynthesis and protein translation. Network N4 illustrates that genes with such biological functions have been intensively investigated for their role in acute leukemias (Pui and
Evans, 2006; Scholar and Calabresi, 1973; Garcia-Manero et al.,
2009; Redner et al., 1999).
Data files:
distance matrix, distance matrix data, network, network data




