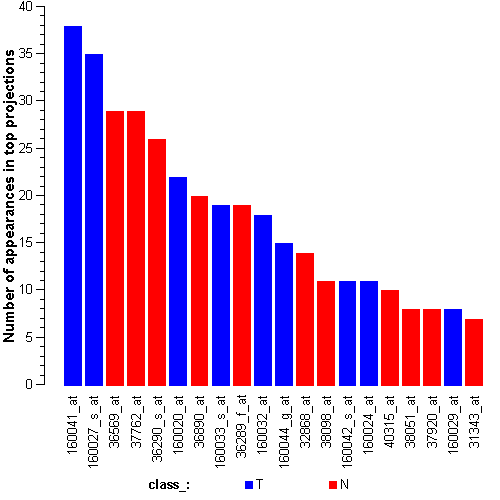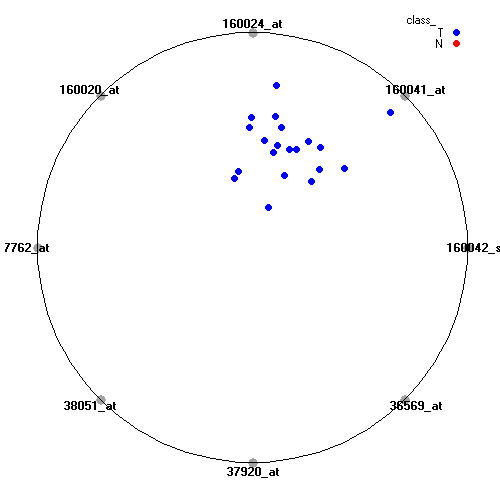
Genes:
160042_s_at: homeo box B6, HOXB6
160041_at: protein tyrosine phosphatase, non-receptor type 18 (brain-derived), PTPN18
160024_at: cyclin-dependent kinase (CDC2-like) 10, CDK10
160020_at: matrix metalloproteinase 14 (membrane-inserted), MMP14
37762_at: epithelial membrane protein 1, EMP1
38051_at: mal, T-cell differentiation protein, MAL
37920_at: paired-like homeodomain transcription factor 1, PITX1
36569_at: C-type lectin domain family 3, member B, CLEC3B
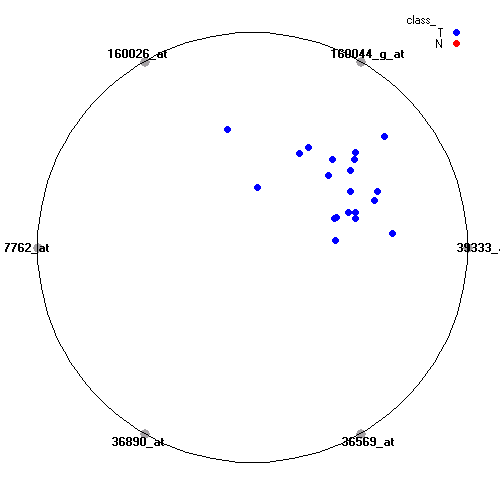
Genes:
39333_at: collagen, type IV, alpha 1, COL4A1
160044_g_at: aconitase 2, mitochondrial, ACO2
160026_at: protein kinase, X-linked, PRKX
37762_at: epithelial membrane protein 1, EMP1
36890_at: periplakin, PPL
36569_at: C-type lectin domain family 3, member B, CLEC3B
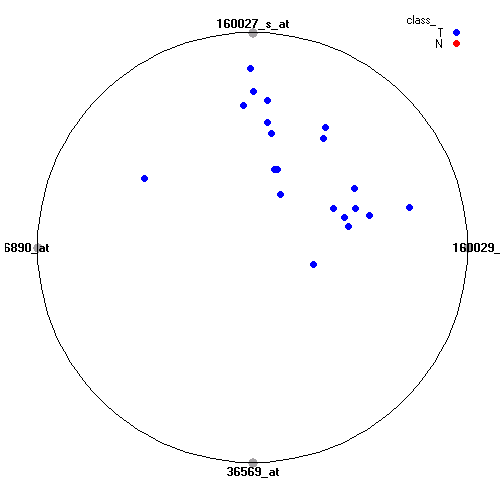
Genes:
160029_at: protein kinase C, beta 1, PRKCB1
160027_s_at: insulin-like growth factor 2 receptor, IGF2R
36890_at: periplakin, PPL
36569_at: C-type lectin domain family 3, member B, CLEC3B