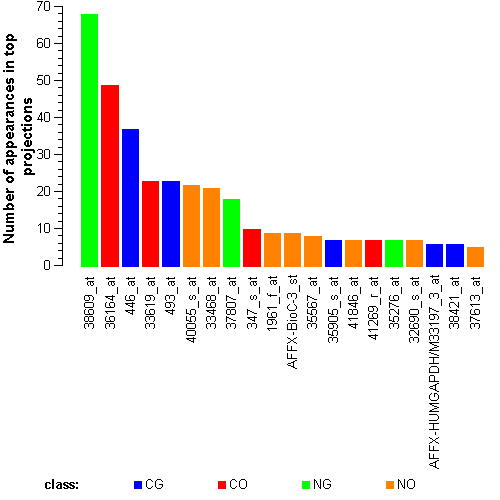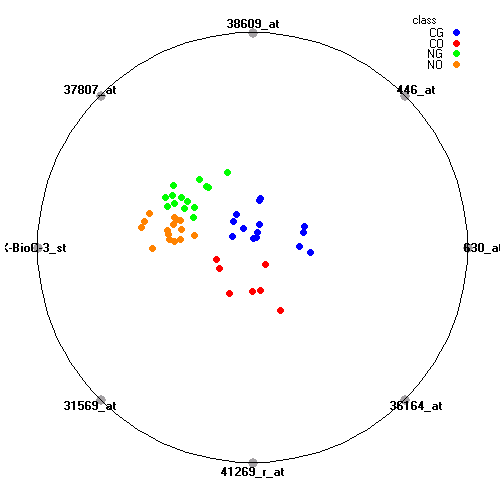
Genes:
630_at: dCMP deaminase, DCTD
446_at: "casein kinase 1, gamma 2", CSNK1G2
38609_at: "sarcoglycan, alpha (50kDa dystrophin-associated glycoprotein)", SGCA
37807_at: "glucokinase (hexokinase 4, maturity onset diabetes of the young 2)", GCK
AFFX-BioC-3_st: ---, ---
31569_at: "solute carrier family 1 (glutamate transporter), member 7", SLC1A7
41269_r_at: apoptosis inhibitor 5, API5
36164_at: "pyruvate dehydrogenase complex, component X", PDHX
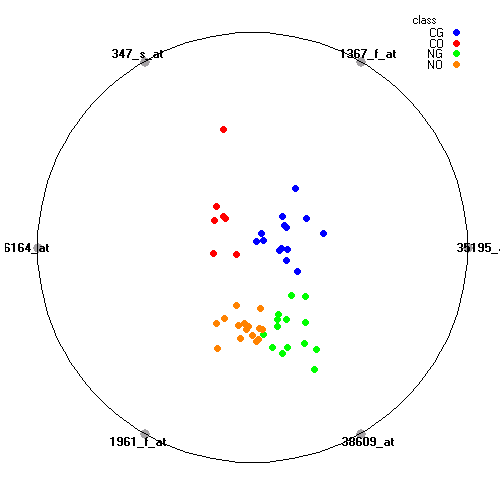
Genes:
35195_at: RNA terminal phosphate cyclase domain 1, RTCD1
1367_f_at: ubiquitin C, UBC
347_s_at: ribosomal protein S23, RPS23
36164_at: "pyruvate dehydrogenase complex, component X", PDHX
1961_f_at: nitric oxide synthase 3 (endothelial cell), NOS3
38609_at: "sarcoglycan, alpha (50kDa dystrophin-associated glycoprotein)", SGCA
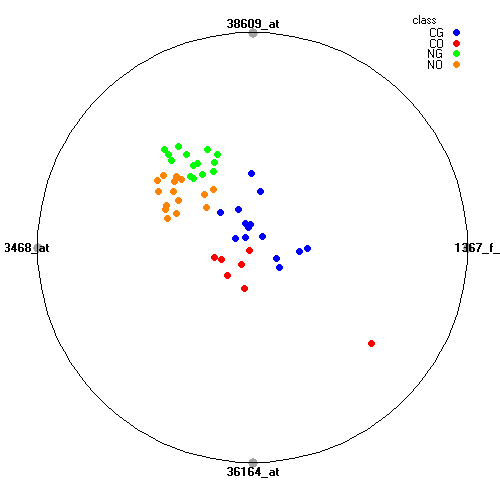
Genes:
1367_f_at: ubiquitin C, UBC
38609_at: "sarcoglycan, alpha (50kDa dystrophin-associated glycoprotein)", SGCA
33468_at: desmoglein 2, DSG2
36164_at: "pyruvate dehydrogenase complex, component X", PDHX