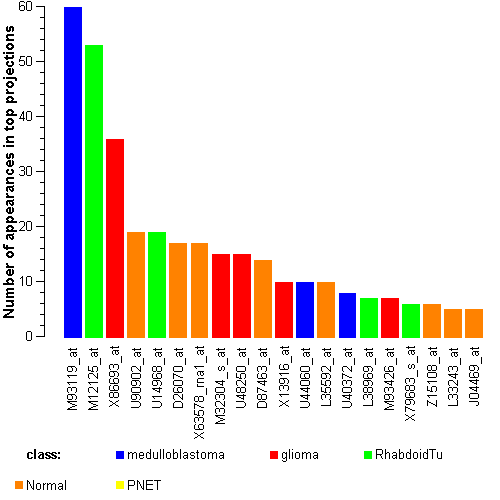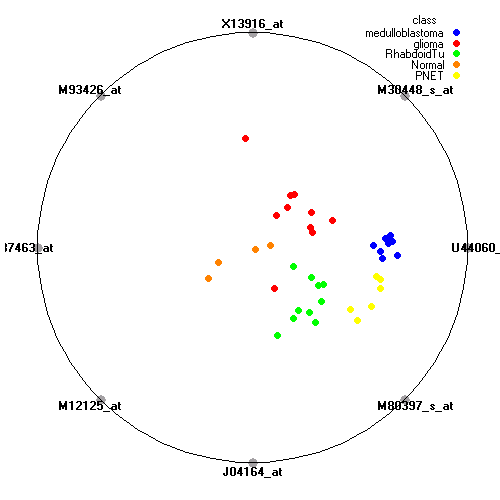
Genes:
U44060_at: prospero-related homeobox 1, PROX1
M30448_s_at: fibrillarin, FBL
X13916_at: low density lipoprotein-related protein 1 (alpha-2-macroglobulin receptor), LRP1
M93426_at: "protein tyrosine phosphatase, receptor-type, Z polypeptide 1", PTPRZ1
D87463_at: phytanoyl-CoA hydroxylase interacting protein, PHYHIP
M12125_at: tropomyosin 2 (beta), TPM2
J04164_at: interferon induced transmembrane protein 1 (9-27), IFITM1
M80397_s_at: "polymerase (DNA directed), delta 1, catalytic subunit 125kDa", POLD1
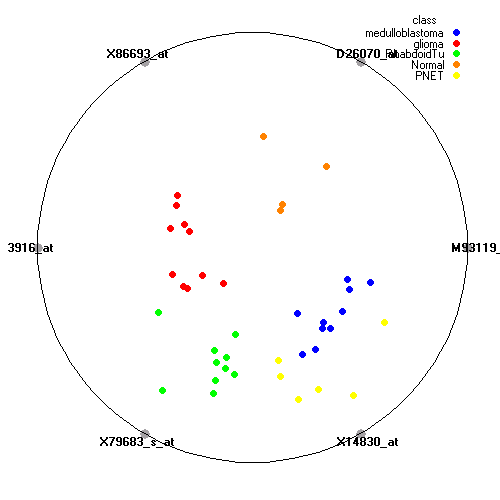
Genes:
M93119_at: insulinoma-associated 1, INSM1
D26070_at: "inositol 1,4,5-triphosphate receptor, type 1", ITPR1
X86693_at: "SPARC-like 1 (mast9, hevin)", SPARCL1
X13916_at: low density lipoprotein-related protein 1 (alpha-2-macroglobulin receptor), LRP1
X79683_s_at: "laminin, beta 2 (laminin S)", LAMB2
X14830_at: "cholinergic receptor, nicotinic, beta polypeptide 1 (muscle)", CHRNB1
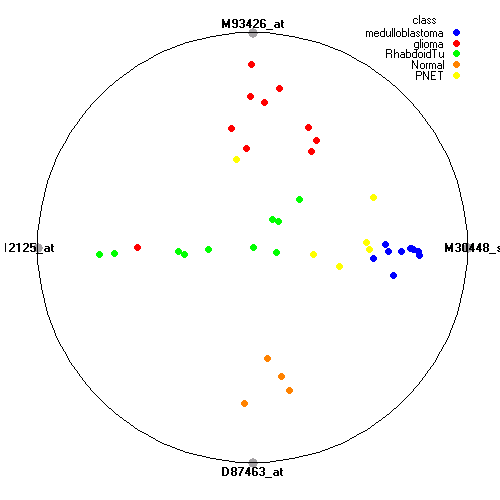
Genes:
M30448_s_at: fibrillarin, FBL
M93426_at: "protein tyrosine phosphatase, receptor-type, Z polypeptide 1", PTPRZ1
M12125_at: tropomyosin 2 (beta), TPM2
D87463_at: phytanoyl-CoA hydroxylase interacting protein, PHYHIP