| FRI > Biolab > Supplements | |||||||||
Data set name: GSE1577Original data set (GSE1577) Data set for Orange
Platform: Affymetrix GeneChip Human Genome U133 Array Set HG-U133A
Number of samples: 19 Note: From the originally measured 22283 probe sets we removed genes that were not present (P) in at least one sample
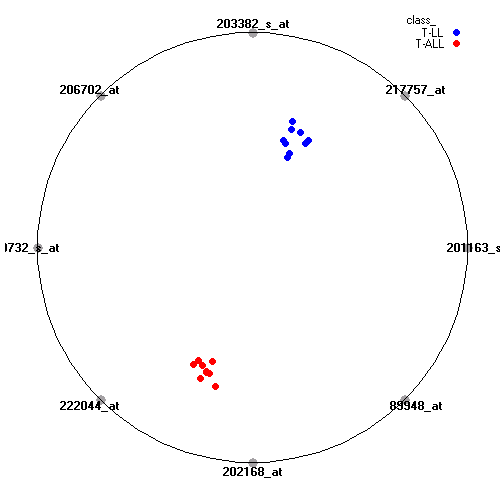
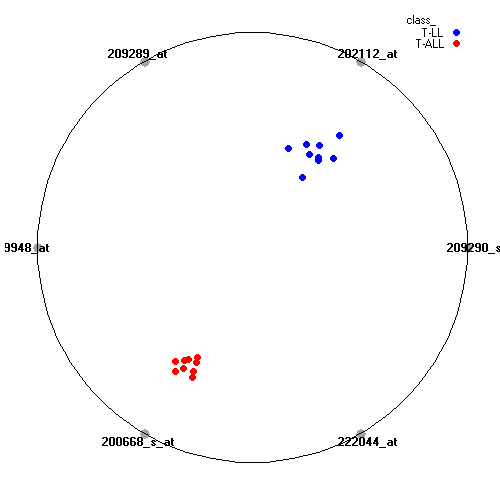
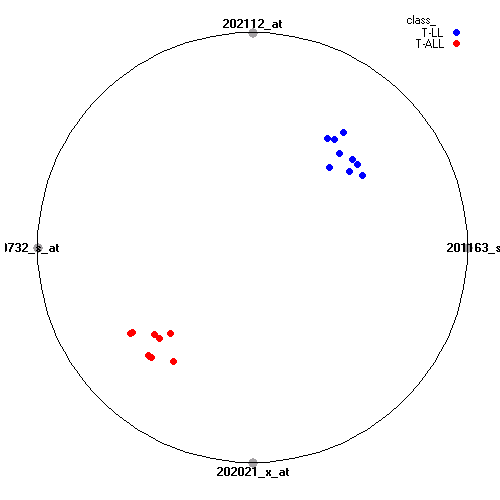
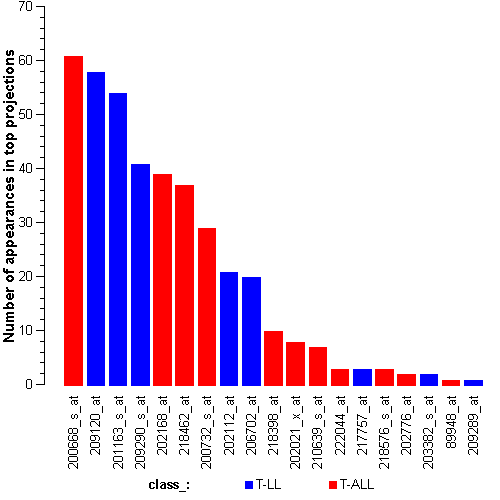
Attribute rankingFollowing is the histogram of genes showing how often are they present in one of the top 100 radviz visualizations with 8 attributes.
|
|||||||||