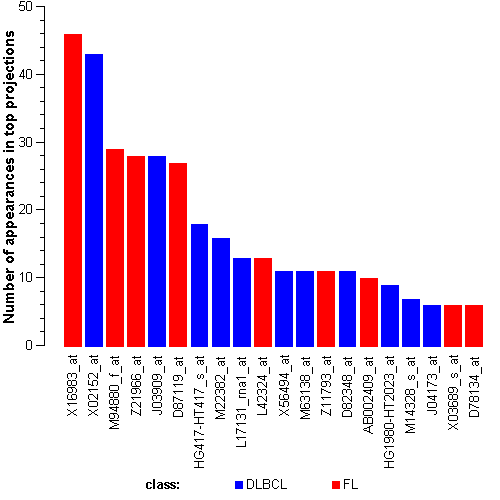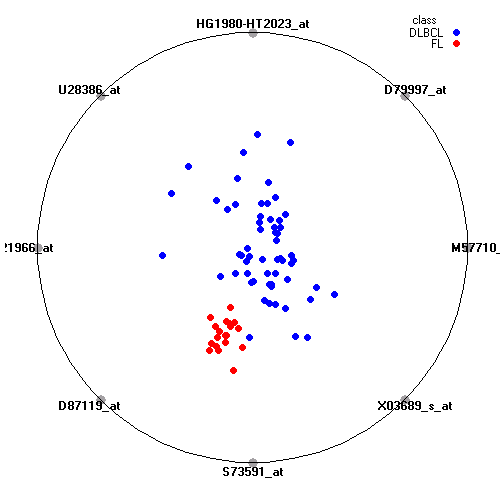
Genes:
M57710_at: "lectin, galactoside-binding, soluble, 3 (galectin 3) /// galectin-3 internal gene", LGALS3 /// GALIG
D79997_at: maternal embryonic leucine zipper kinase, MELK
HG1980-HT2023_at: ---, ---
U28386_at: "karyopherin alpha 2 (RAG cohort 1, importin alpha 1)", KPNA2
Z21966_at: "POU domain, class 6, transcription factor 1", POU6F1
D87119_at: tribbles homolog 2 (Drosophila), TRIB2
S73591_at: thioredoxin interacting protein, TXNIP
X03689_s_at: eukaryotic translation elongation factor 1 alpha 1, EEF1A1
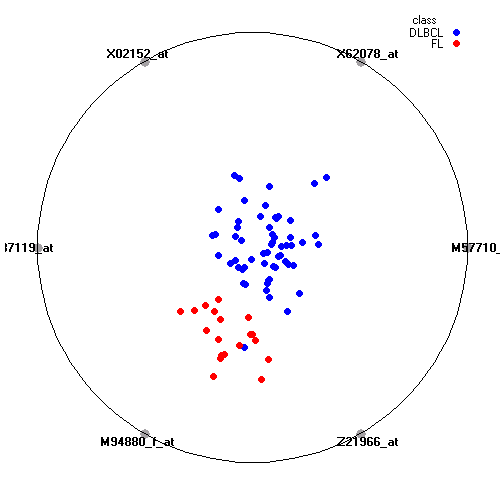
Genes:
M57710_at: "lectin, galactoside-binding, soluble, 3 (galectin 3) /// galectin-3 internal gene", LGALS3 /// GALIG
X62078_at: GM2 ganglioside activator, GM2A
X02152_at: lactate dehydrogenase A, LDHA
D87119_at: tribbles homolog 2 (Drosophila), TRIB2
M94880_f_at: "major histocompatibility complex, class I, A", HLA-A
Z21966_at: "POU domain, class 6, transcription factor 1", POU6F1
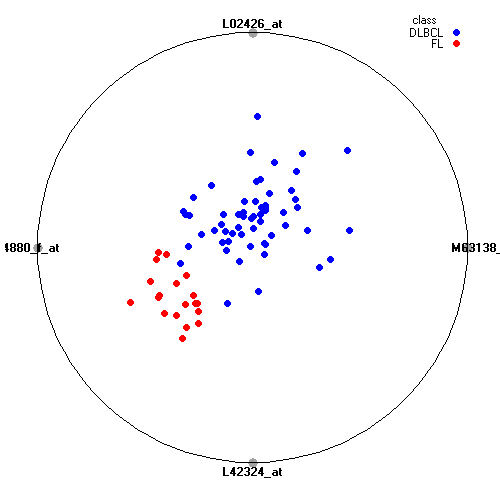
Genes:
M63138_at: cathepsin D (lysosomal aspartyl peptidase), CTSD
L02426_at: "proteasome (prosome, macropain) 26S subunit, ATPase, 1", PSMC1
M94880_f_at: "major histocompatibility complex, class I, A", HLA-A
L42324_at: G protein-coupled receptor 18, GPR18